Calls for Ukraine
Calls for Europe
Calls for USA
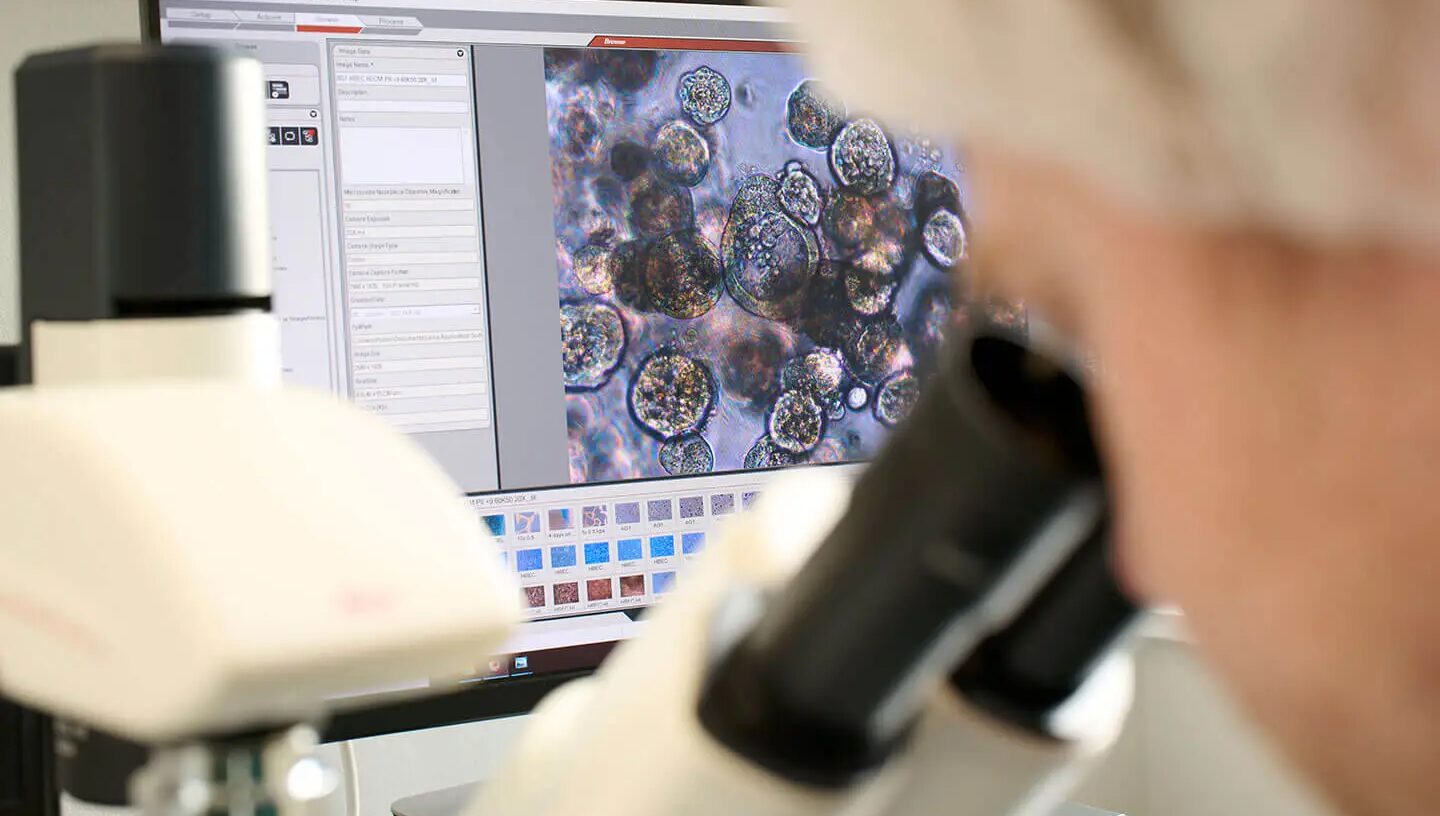
American researchers have developed new software that, similar to meteorological models, allows predicting the behavior of cells in tissues over time. This technology combines genomic data and mathematical modeling, paving the way for the creation of personalized “digital twins” of patients, which will ultimately help determine the best treatment strategy. The study was published in the journal Cell.
Traditional methods of monitoring the condition of cancer patients, especially those using modern genomic technologies, provide only a “snapshot” of the tumor’s condition at a given moment in time. They do not show how cells communicate with each other and do not reflect the real dynamics of such interactions, which lead to tumor growth or resistance to treatment. It is especially difficult to predict the behavior of cancer due to the individual characteristics of each patient’s immune system and the complex tumor ecosystem, which includes not only tumor cells but also many healthy cells. Until now, scientists lacked a tool that could show how a tumor behaves dynamically — how interactions between cells change and how this affects the effectiveness of treatment.
The most significant achievement of the new project was the development by a group of researchers of the so-called “grammar of hypotheses” — a system that allows the behavior of cells (movement, division, interaction with other cells and the immune system) to be described using simple sentences in English. Based on this, scientists have created digital models of multicellular biological systems and facilitated interaction between specialists from different fields. “This grammar helps not only to link biology with code, but also to simplify interdisciplinary collaboration,” explained Daniel Bergman of the University of Maryland. The authors of the work combined this grammar with spatial transcriptomic data from real tumor samples, which allow determining which genes are active and where exactly in the tissue. This made it possible to build complex computational models that simulate tumor development over time.
The researchers applied the model to two different types of cancer. In breast cancer, they were able to show how a malfunction of immune cells (T lymphocytes) can contribute to the spread of cancer cells from the primary tumor site to neighboring tissues. In the second case, the specialists used data from patients with pancreatic cancer to simulate the response to immunotherapy. The model predicted that the response to treatment would vary among each “virtual” patient, highlighting the importance of the tumor’s cellular ecosystem. For example, fibroblasts — healthy cells that form a dense structure around the tumor — actively interact with cancer cells and influence their responses. Spatial transcriptomics allowed these interactions to be visualized, and the model tracked their impact on tumor growth and invasion. “Immune cells are amazing and obey rules of behavior that can be recreated in a model,” said Jeanette Johnson of the University of Maryland. “This system allows us to freely explore hypotheses about what happens in a patient’s body over time, without cost or risk to the patient.”
The study paves the way for the creation of a new virtual platform that can be used to model the course of disease in a specific patient and test the effectiveness of different treatment options in a simulated environment. This method makes it possible to conduct “virtual clinical trials” and promote a personalized approach to cancer treatment. Its versatility is confirmed by the fact that the model has also been successfully applied in neuroscience: a team from Johns Hopkins University used the same principle to model the formation of brain layers. “Ever since I moved from weather forecasting to computational biology, I believed that we could apply the same principles to oncology,” said lead author Elana J. Fertig of the University of Maryland. “Adapting this approach to genomic technologies gives us a virtual laboratory where we can conduct experiments entirely in silico.” The developed grammar is already available to the scientific community as open source, which will contribute to its further development. Scientists hope that such models will eventually become an indispensable tool in the fight against cancer.
Please rate the work of MedTour
